SnapGene Updates
The latest SnapGene version is 7.2.0.
Update to SnapGene 7
Subscribers
Update for free by downloading the installer for your system (instructions):
Perpetual Customers
Contact us at support.snapgene.com to learn about upgrade options.
Release Notes
SnapGene 7.2 provides a new visualization of primer homodimer structures and enhancements to file management, allowing tabs to be organized in multiple windows using drag and drop, and improvements to interacting with files in projects.
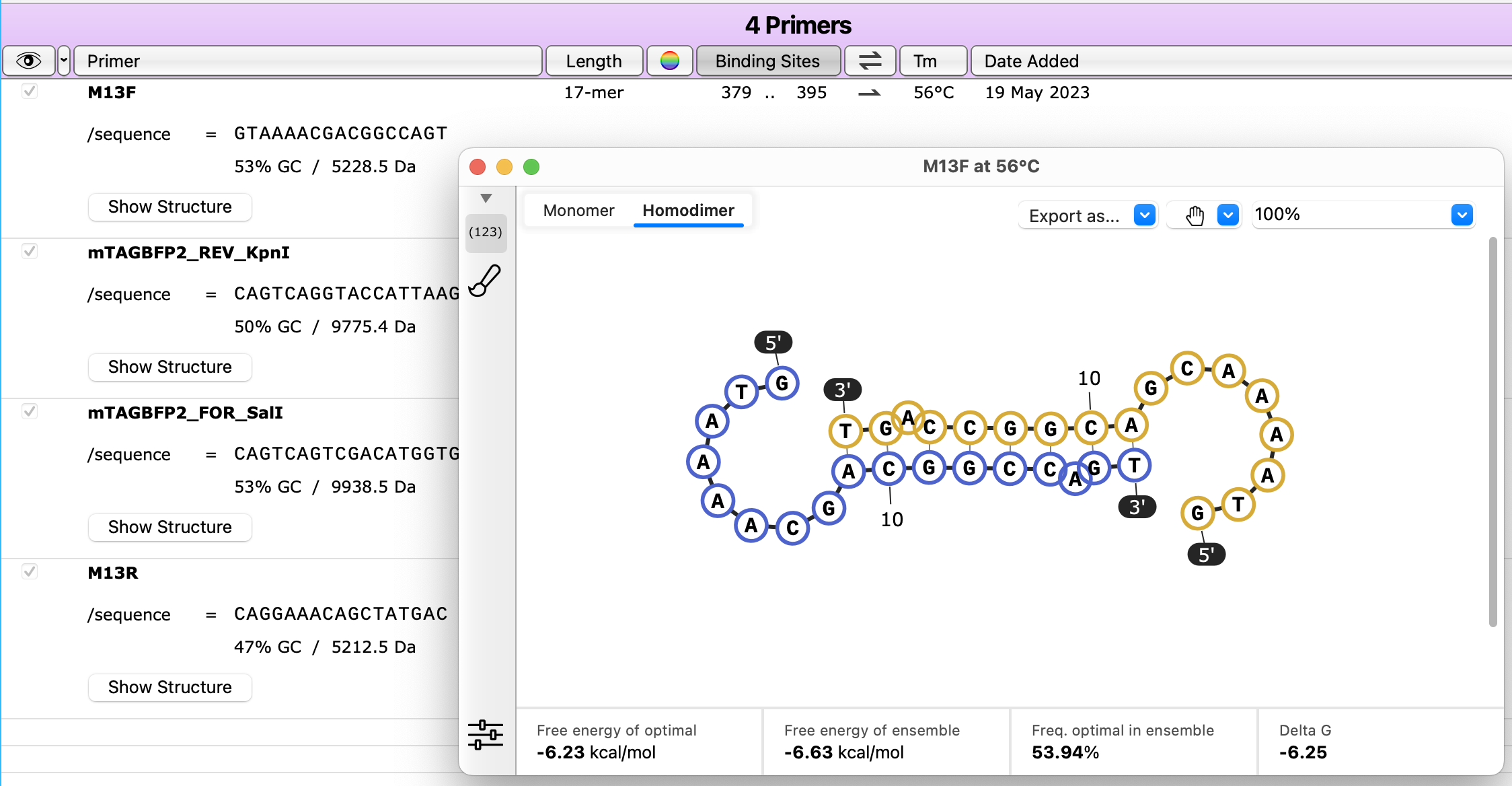
Calculate Primer Homodimer Structures
Check your primers for self-dimerization by viewing homodimer structures with deltaG values, via the Show Structure buttons in Primers view.
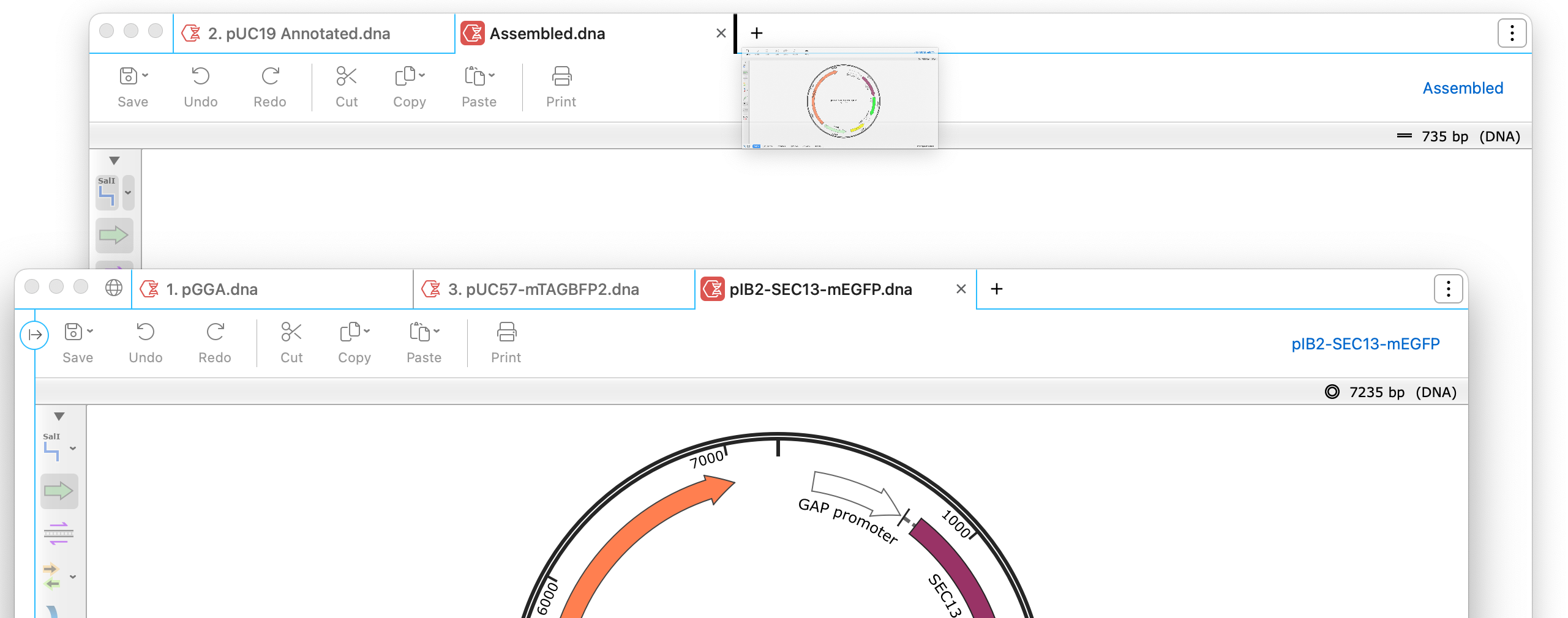
Draggable File Tabs
Easily group your sequences by dragging file tabs in and out of the window to create multiple tabbed windows.
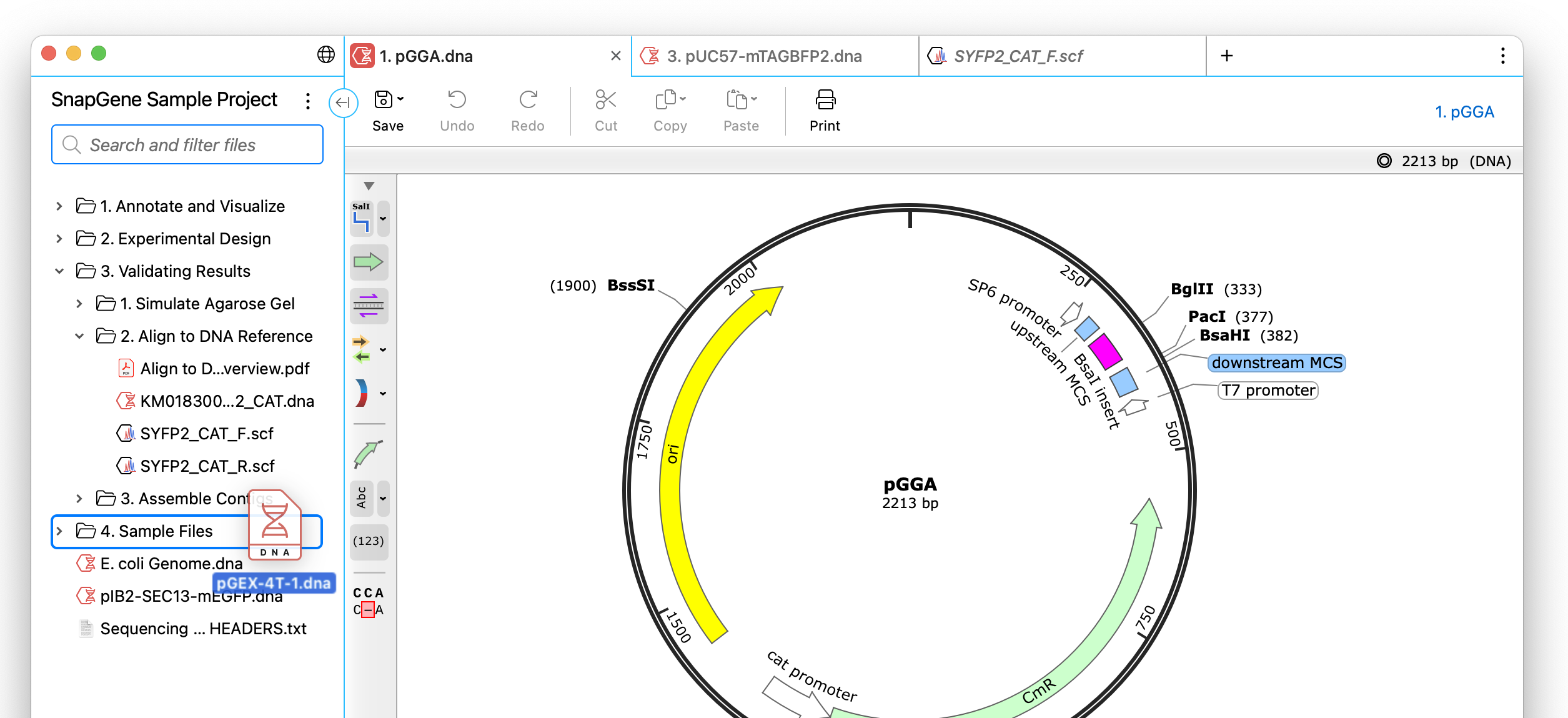
UX Enhancements for Interacting with Files
Smoother importing and interacting with tabs, files, and folders to address common issues, including enhancing the visibility of the current tab and file name, improved default view settings for chromatograms, drag and drop import into folders, and more.
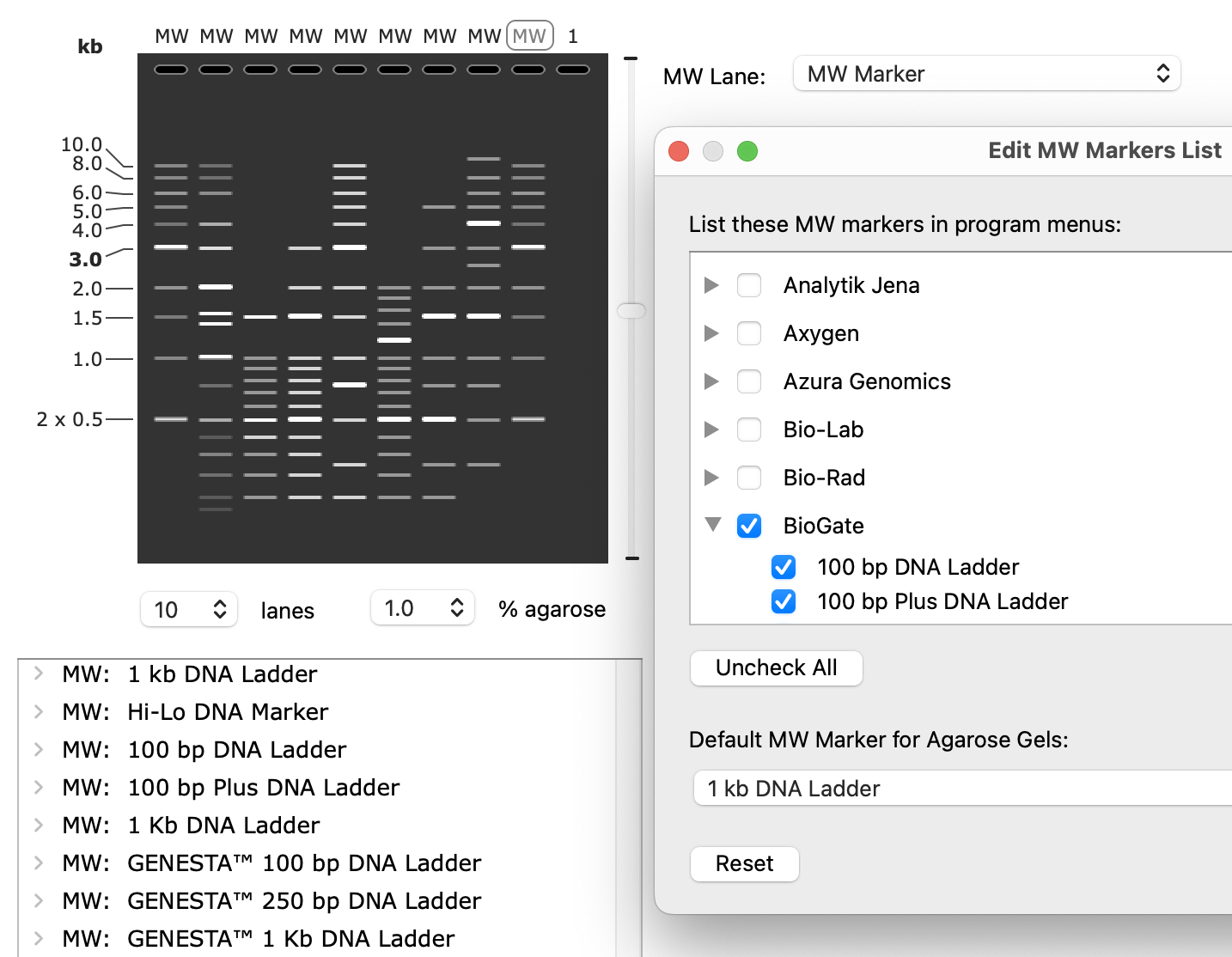
New Common Features and Agarose Gel Ladders
Can now choose gel ladders from several additional suppliers, and more features have been added to the standard feature database for detection in your sequences.
This release includes a number of important fixes and improves overall stability.
SnapGene 7.1 provides enhancements to streamline file search within a selected folder, introduces secondary structure visualizations for ssDNA sequences including primers, and includes a variety of fixes to improve stability, file import workflows and other functions.
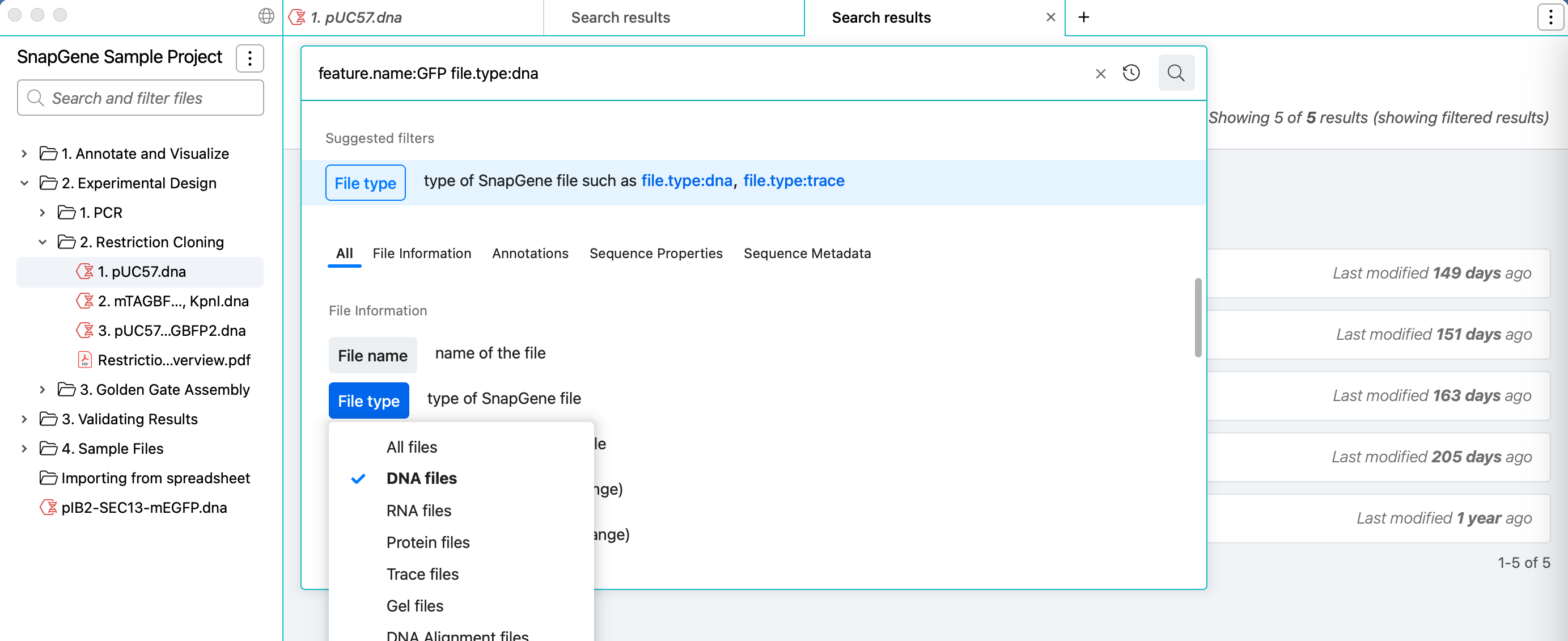
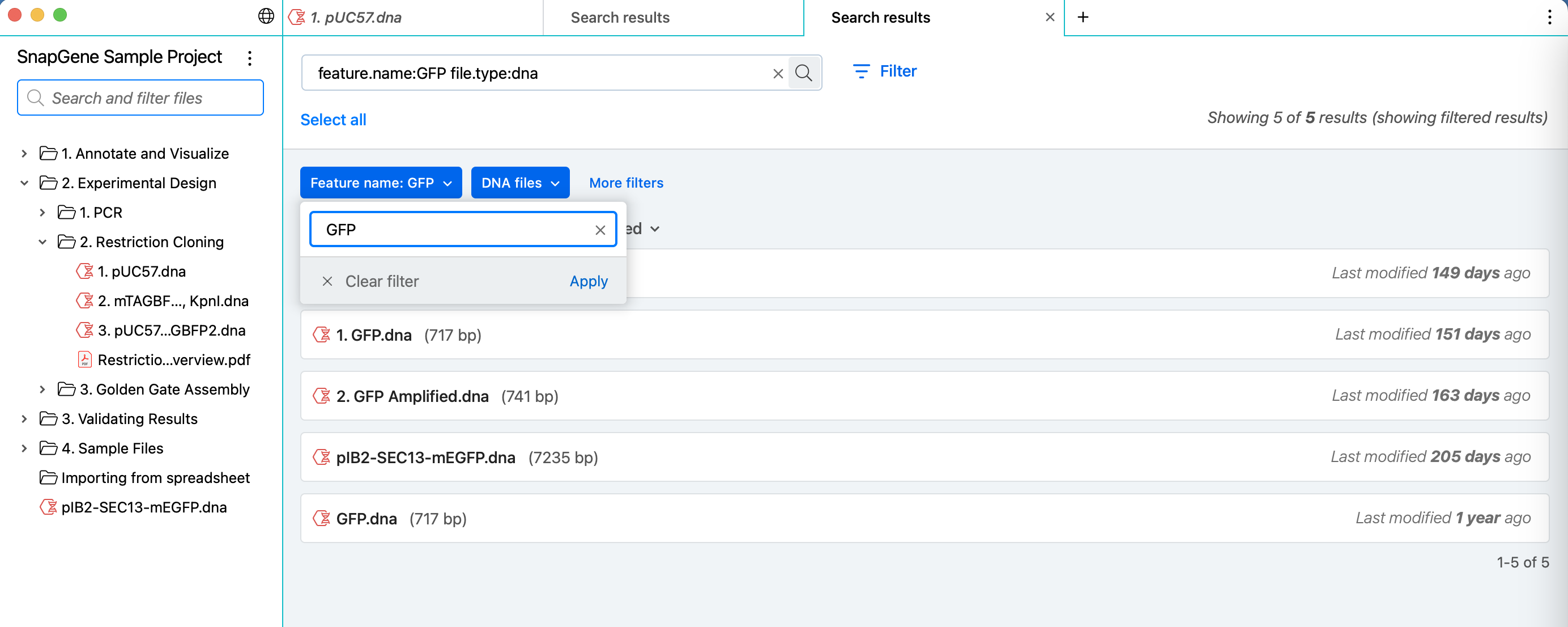
File Search Enhancements
Quickly and easily build search queries to find your sequences and other files in the selected project folder with simplified filter entry controls, and run multiple searches in different tabs.
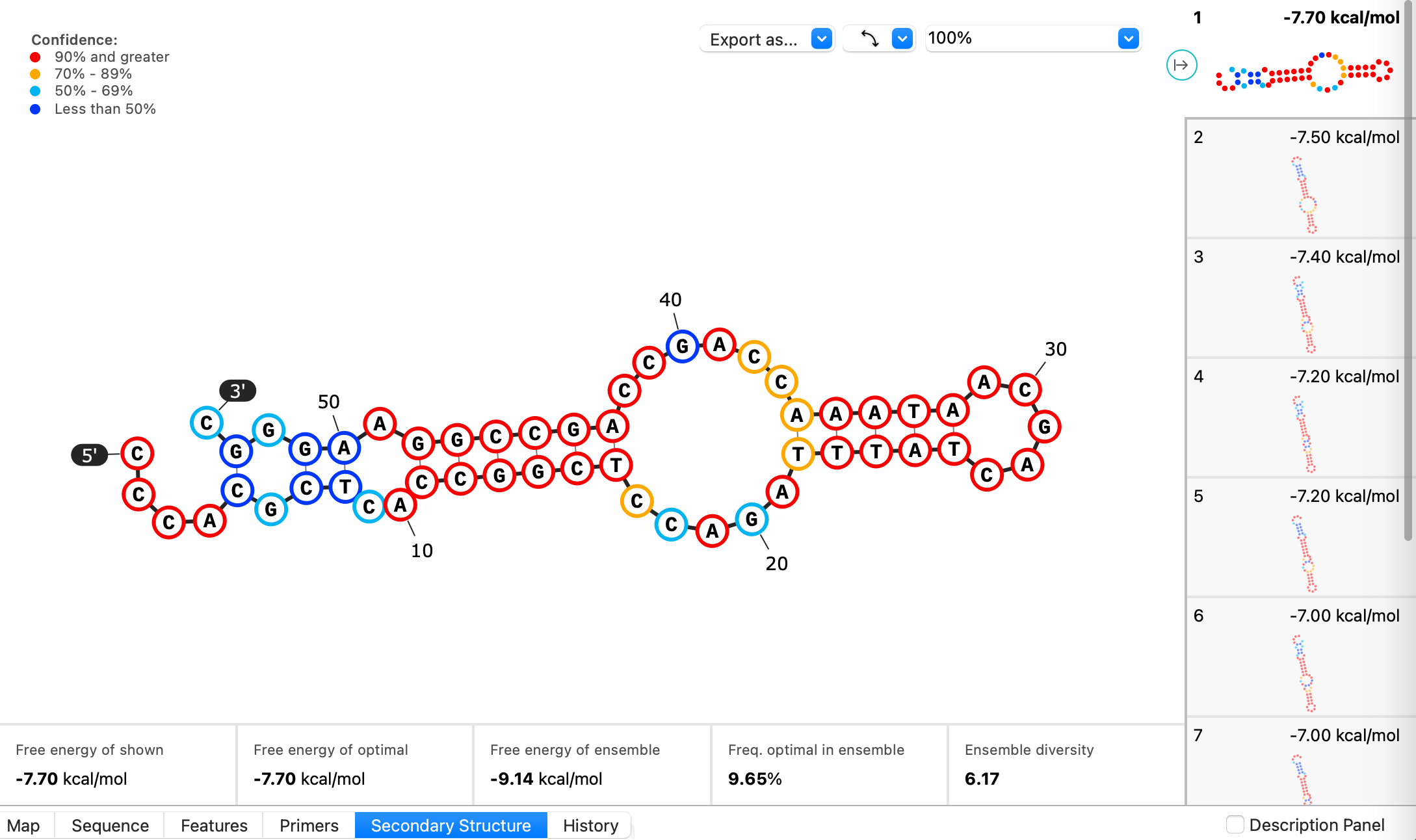
Secondary Structure View for Single-Stranded DNA Sequences
Visualize the secondary structure for single-stranded DNA sequences calculated with the ViennaRNA package via the new secondary structure tab
View Primer Hairpins and Secondary Structures
Check your primers for hairpins or other self-binding secondary structures with a new button for calculating secondary structures for primer monomers, available in the primers tab.
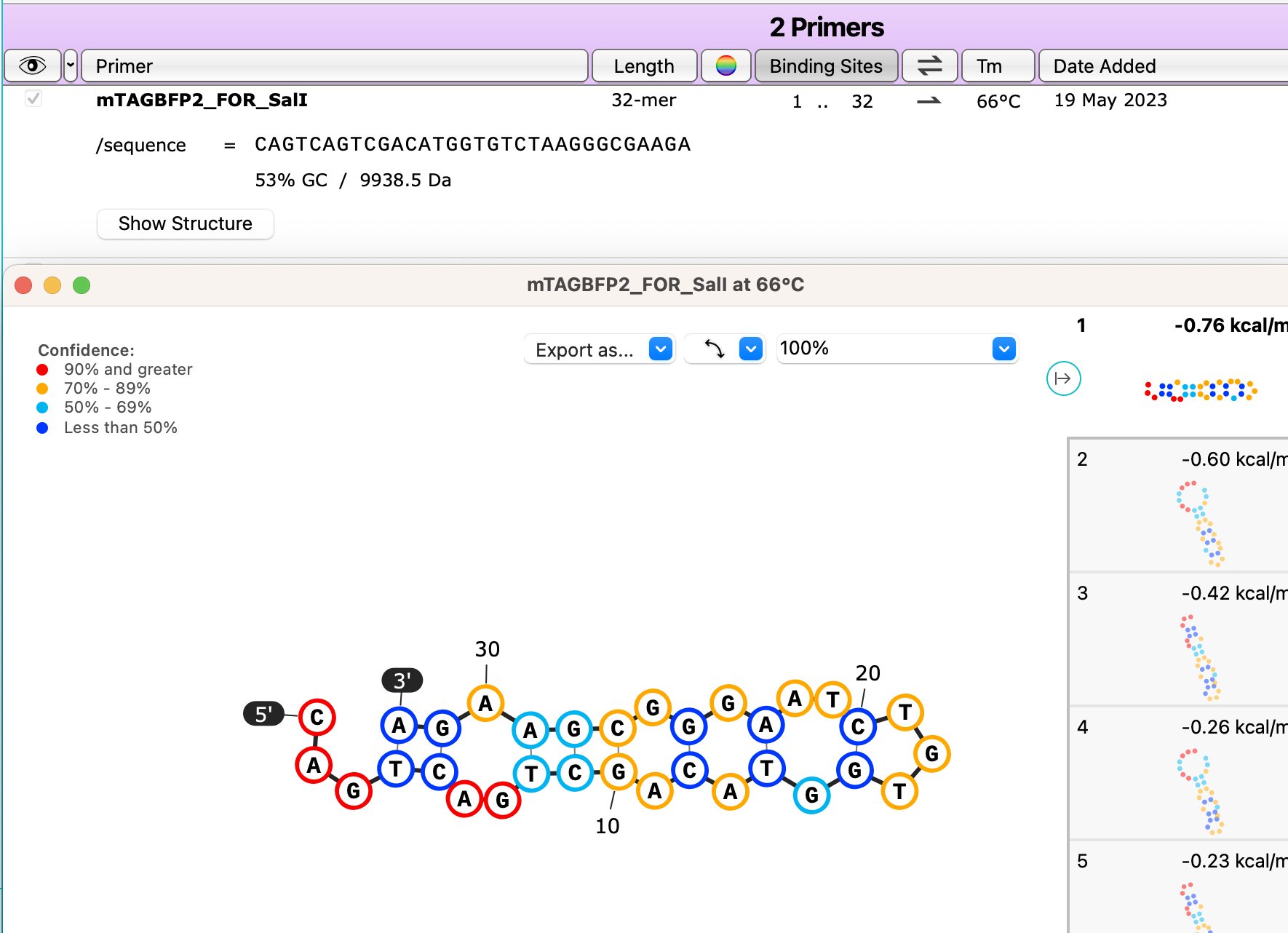
This release includes a number of stability and performance improvements as well as other fixes.
This release includes a number of important fixes and improves overall stability.
This release includes a number of important fixes and improves overall stability.
SnapGene 7.0 introduces an updated look and feel with enhanced data management capabilities. Easily navigate files and folders with tabs, and perform sequence and metadata-based searches for files directly from the new folder panel. Other enhancements include new primer insertion annotations and various usability improvements.
Tabbed Interface
Navigate between documents using the new tabbed interface, or pop out documents into separate windows for comparison purposes.
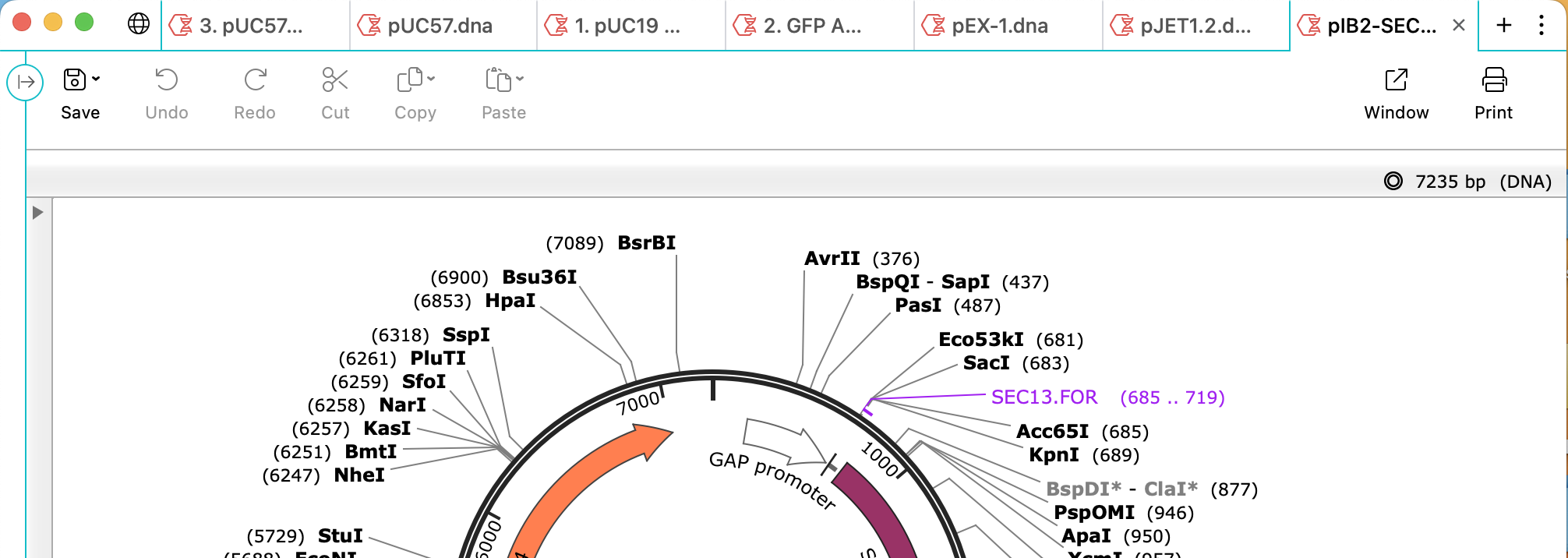
File and Folder Management
Choose any folder on your file system as a project and quickly view subfolders and files directly within a new, collapsible, folder panel. Perform standard file operations and quickly browse all supported file types.
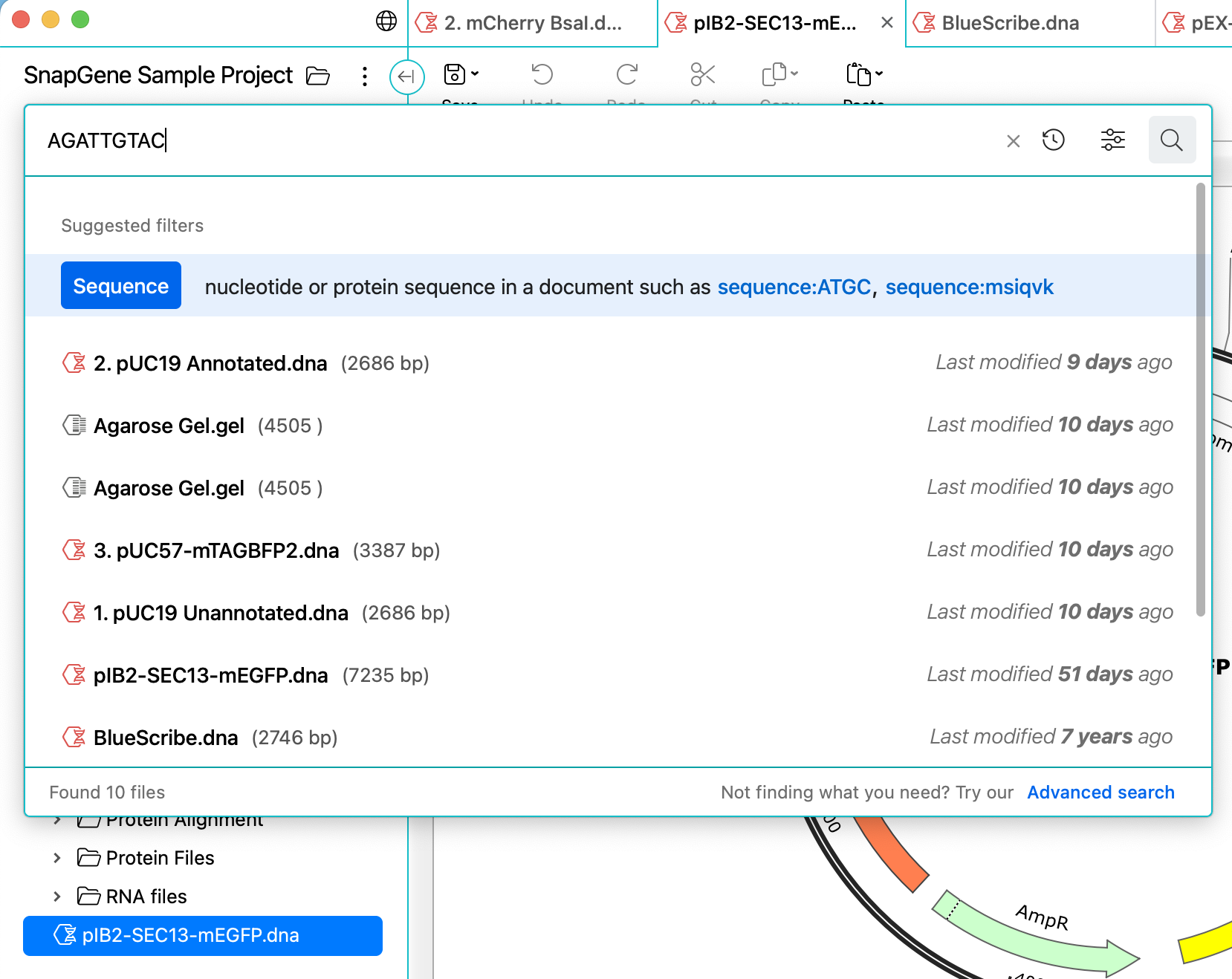
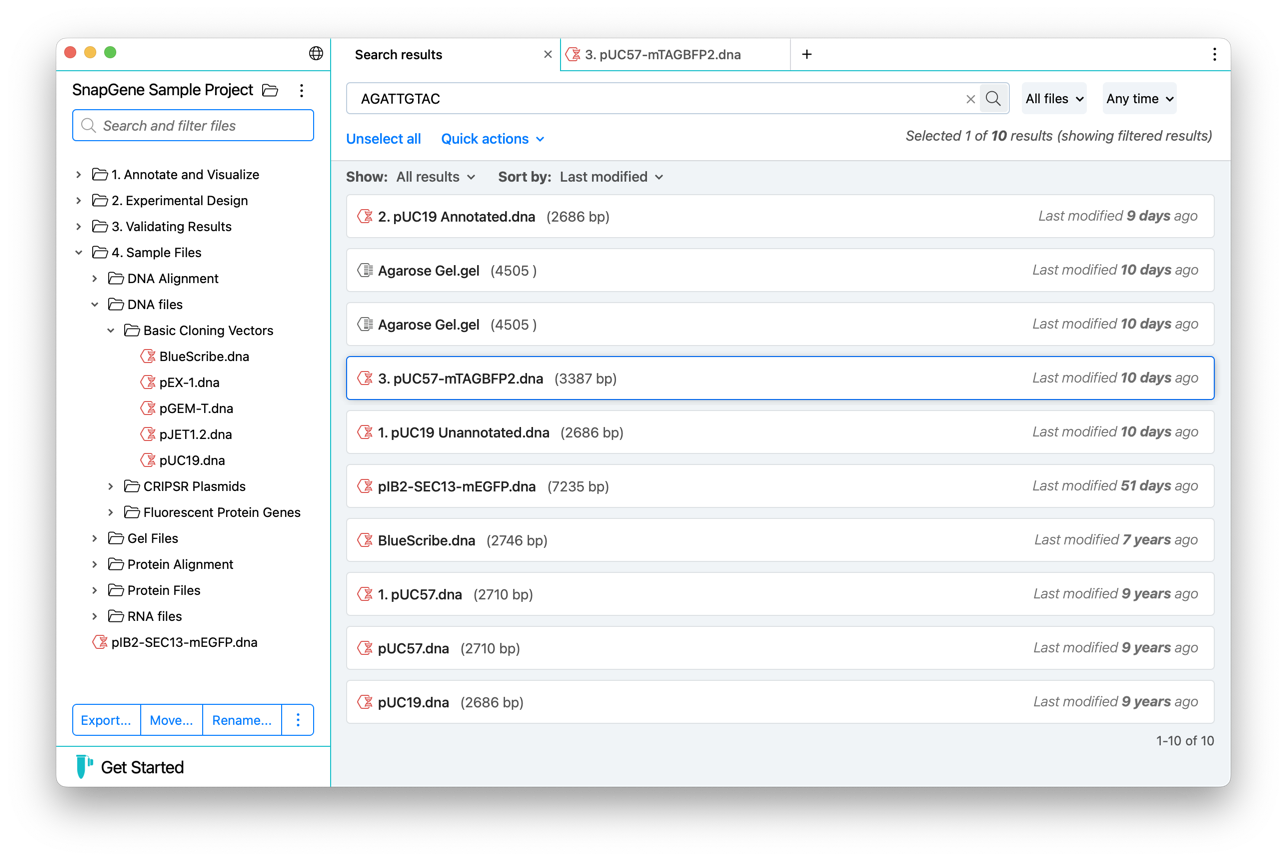
File Search
Quickly find files within a project using attributes such as the name or type, containing sequence, feature annotations and more via a flexible, modern interface.
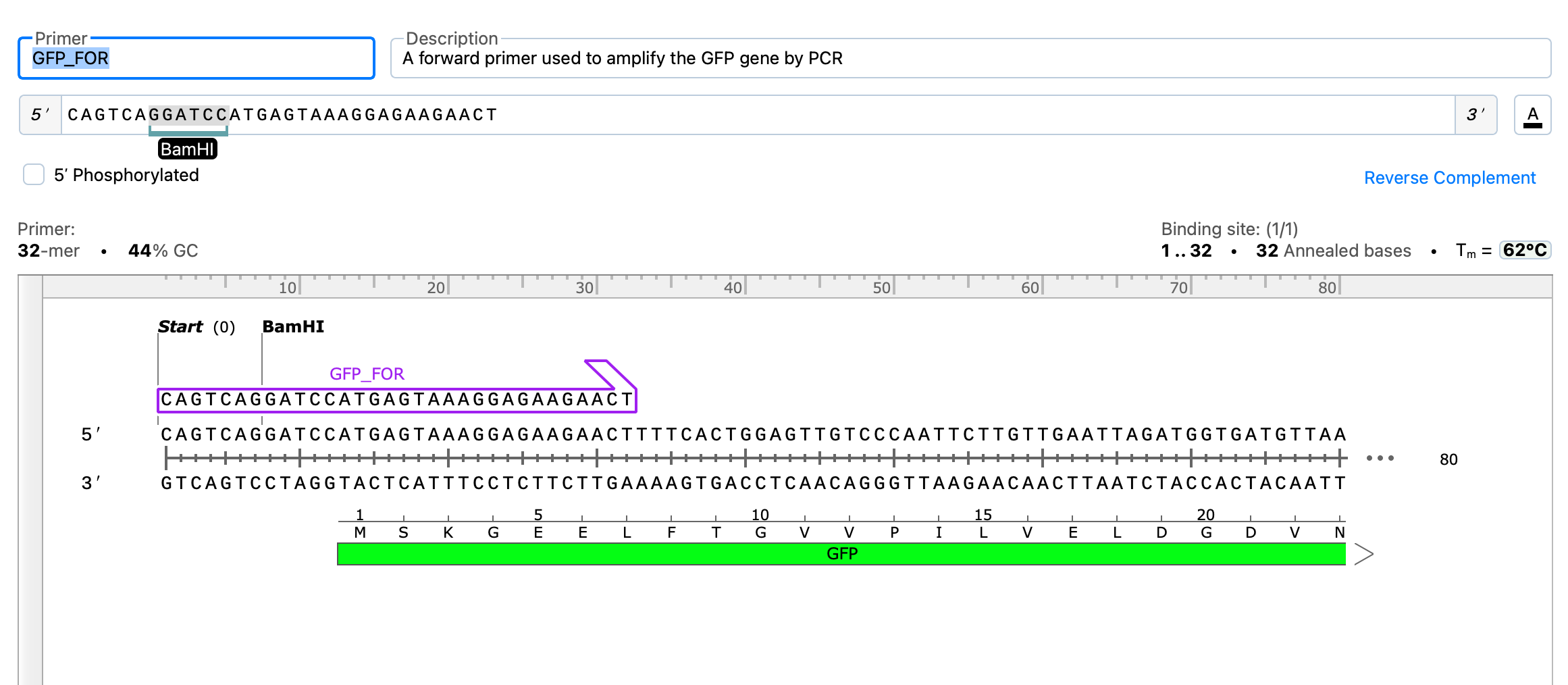
Primer Annotations
Visualize insertions such as enzymes, codon or short peptide sequences added to your primers in the updated Add/Edit Primer dialog. Regions of interest (Golden Gate restriction sites, Gateway cloning attB sites, directional TOPO motif) are now annotated for primers designed automatically as well.
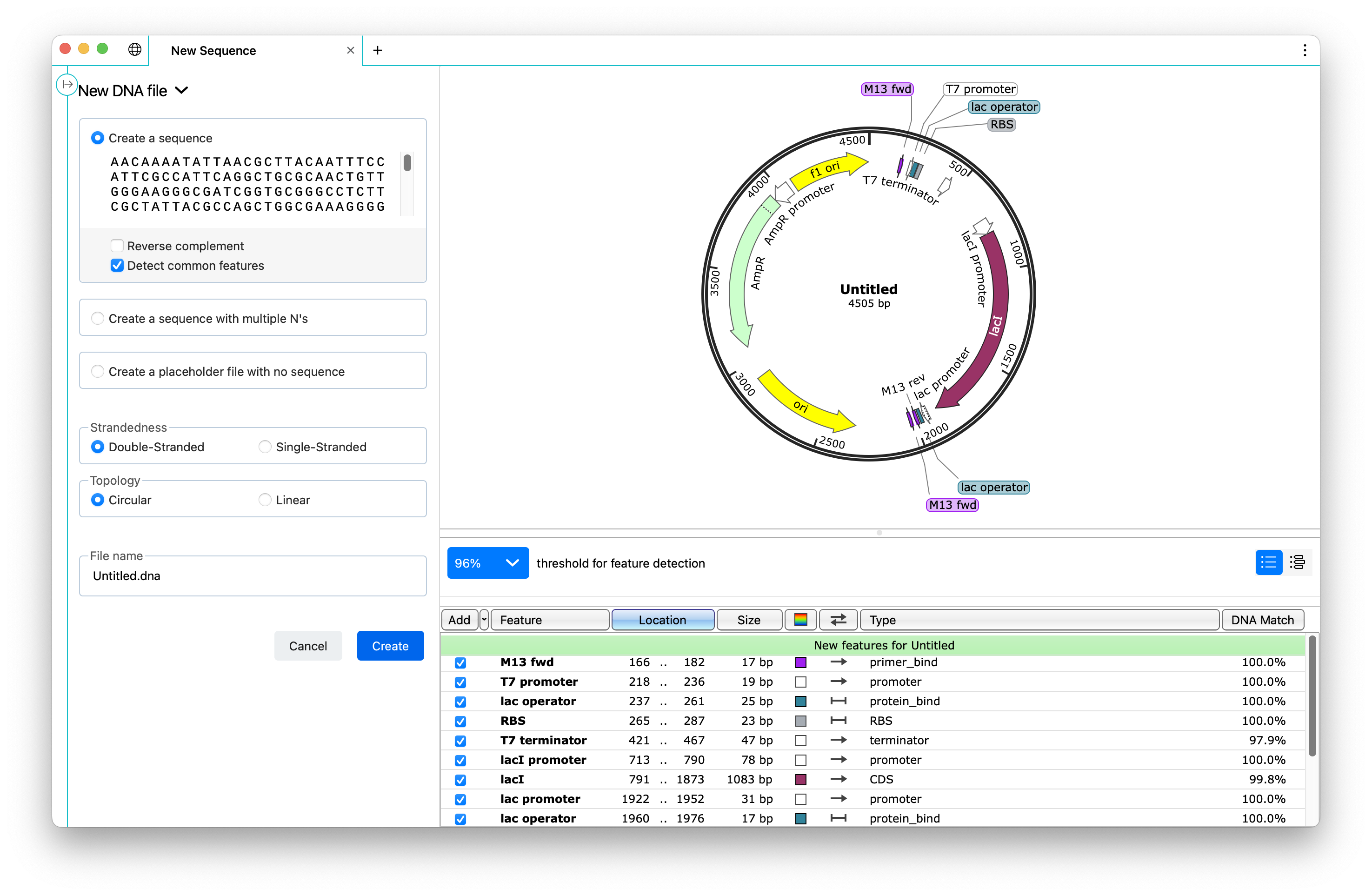
Preview Detected Features for New Sequences
The new file pane now includes a sequence map to preview features that are detected in your sequence and allows you to choose which annotations to include in your sequence.
Get Started
Quickly access recent files and folders using the new Get Started pane, or learn more about how to use SnapGene with links to short video tutorials, user guide articles, and the SnapGene Academy.

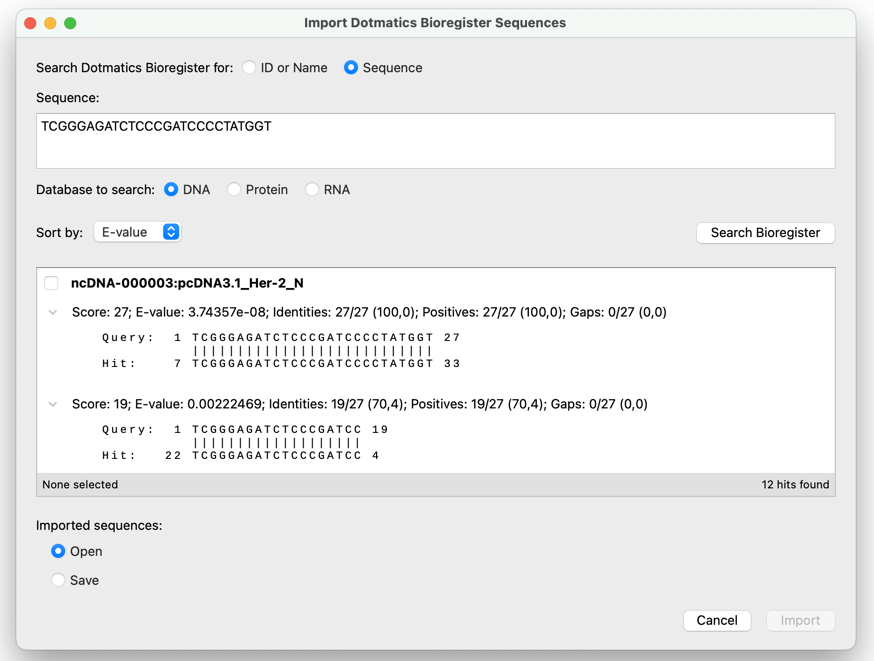
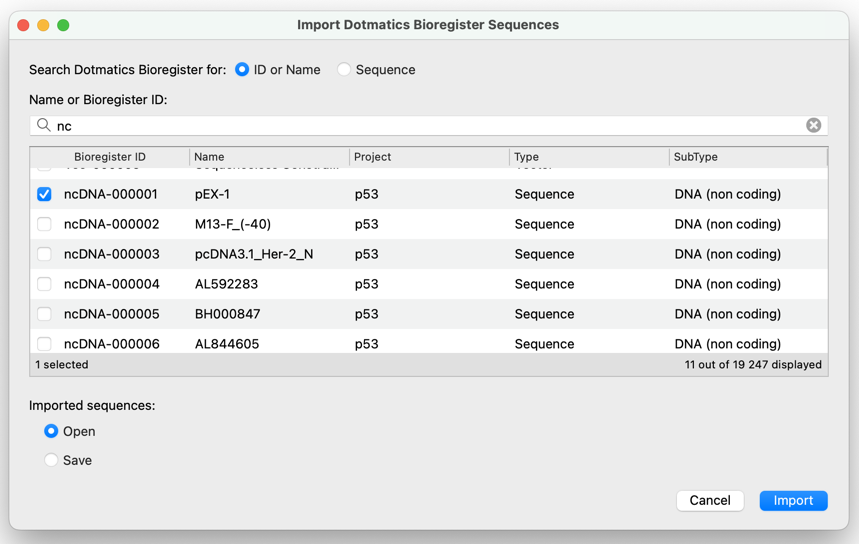
Search for and Import Sequences from Dotmatics Bioregister
Quickly and easily search for sequences in Dotmatics Bioregister directly from within SnapGene based on the sequence name, Bioregister ID, or a similar sequence (BLAST search). Save a sequence of interest locally or open it in SnapGene right away. This integration requires Bioregister 6.1.0 or later.
Version 6.2.1. enables registering sequences in Dotmatics Bioregister in addition to a fixing various other issues.
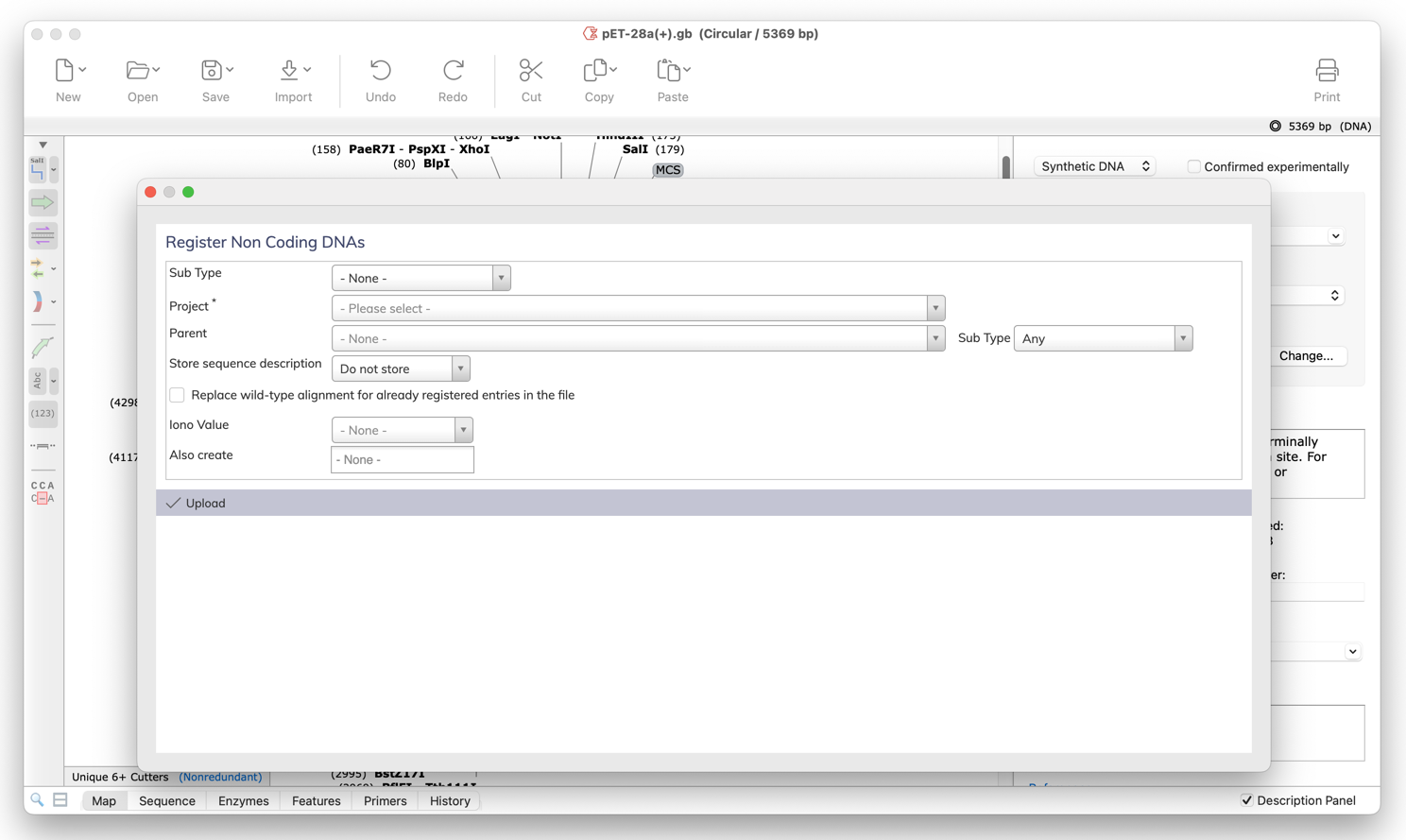
Register in Dotmatics Bioregister
Quickly and easily register your sequences in Dotmatics Bioregister directly from within SnapGene. It is no longer necessary to first save using a specific file format and manually upload to Bioregister using a web browser. This integration requires Bioregister 22.2 or later.
Version 6.2 adds support for suboptimal RNA secondary structures and includes a number of other enhancements to the Structure view, includes a number of improvements to the Golden Gate cloning simulation tools and PCR simulations in general, provides better flexibility when working with Protein properties, and modernized buttons and icons.
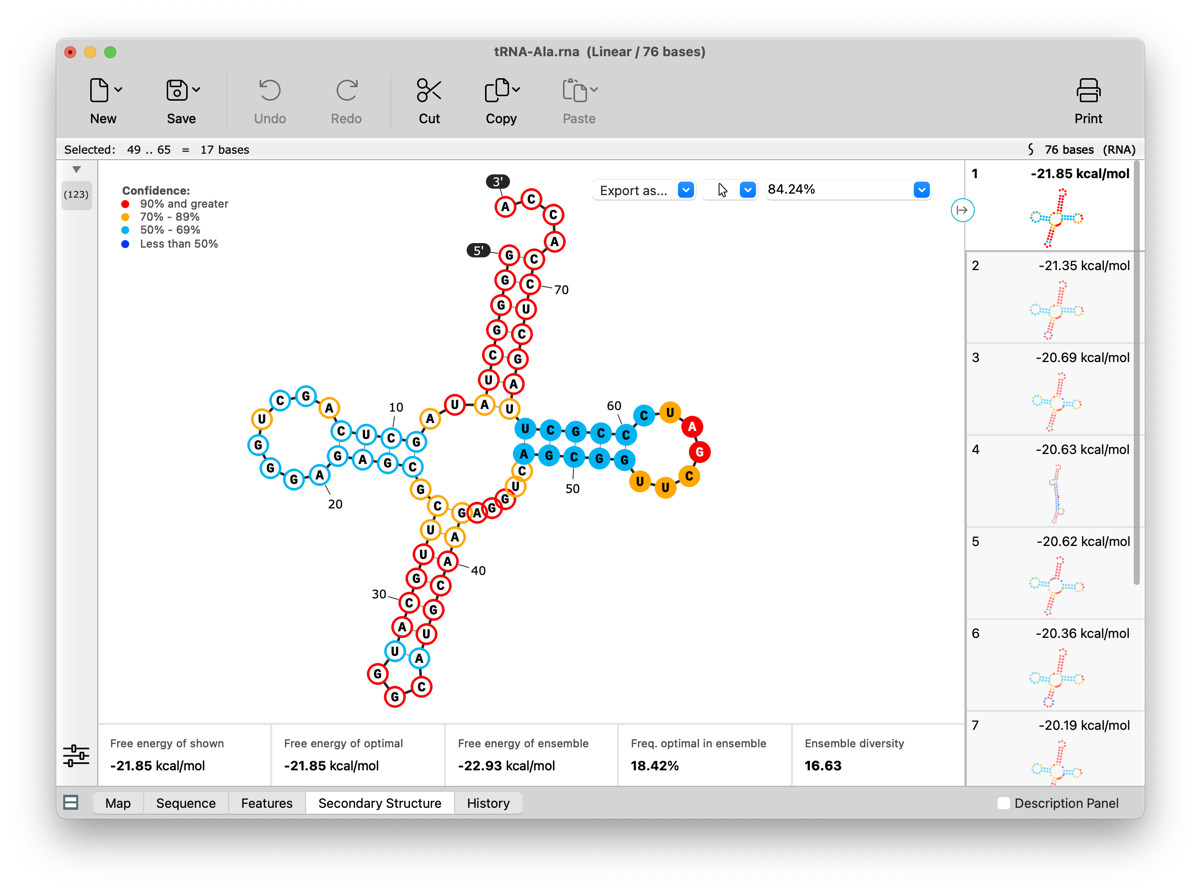
RNA Secondary Structure view enhancements
Suboptimal RNA secondary structures are now shown. All structures can be recalculated using adjusted Tm and other settings. Visualization and selection capabilities have been enhanced, including the ability to display and make sequence selections that are synced across all views. Coordinates and 5' / 3' end labels can optionally be displayed. See View Predicted RNA Secondary Structure for more information.
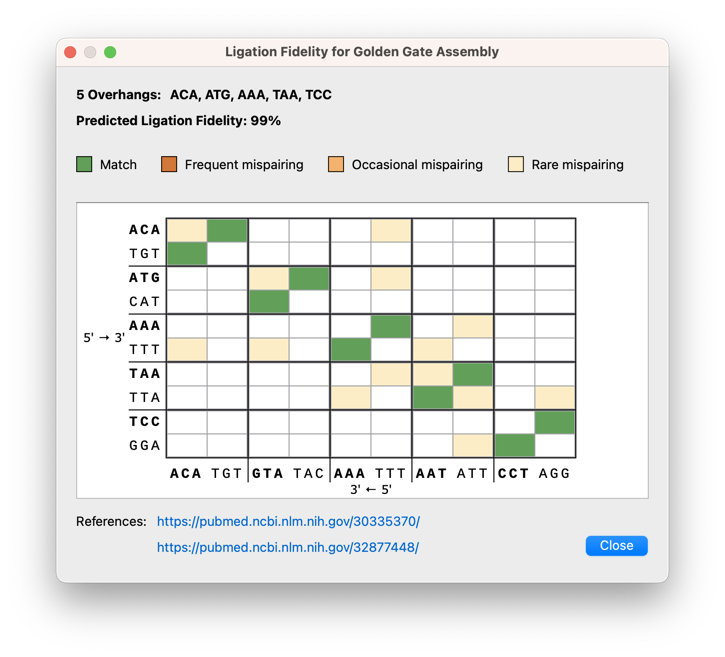
Golden Gate Assembly Improvements
Various enhancements to the Golden Gate cloning dialogs include a ligation fidelity matrix for assessing overall reaction fidelity, it is now possible to adjust overhangs for manually designed primers, a new option to cut the vector with any enzymes (not just Type IIS enzymes), as well as an updated settings dialog with easy access to recommended enzymes.
Improvements to primer design in PCR cloning simulations
You can now adjust the hybridization region for all PCR cloning simulations (e.g. PCR, Golden Gate, Gibson, In Fusion, etc.). When using automatic primer design the hybridized region will be configured automatically, ensuring miscellaneous features are not transferred to the product when a 5’ primer extension by chance partially hybridizes to the template.
Protein properties can now be copied and exported
Copy or export the properties and amino acid data for full protein sequences or selected regions in text or spreadsheet (*.csv / *.tsv) formats. Individual properties can also be copied directly from the view.
Modernized toolbar, application, and document icons
SnapGene now uses modernized icons in the top toolbar. The application icon and file icons have also been updated.
Version 6.2 adds support for suboptimal RNA secondary structures and includes a number of other enhancements to the Structure view, includes a number of improvements to the Golden Gate cloning simulation tools and PCR simulations in general, provides better flexibility when working with Protein properties, and modernized buttons and icons.
RNA Secondary Structure view enhancements
Suboptimal RNA secondary structures are now shown. All structures can be recalculated using adjusted Tm and other settings. Visualization and selection capabilities have been enhanced, including the ability to display and make sequence selections that are synced across all views. Coordinates and 5' / 3' end labels can optionally be displayed. See View Predicted RNA Secondary Structure for more information.
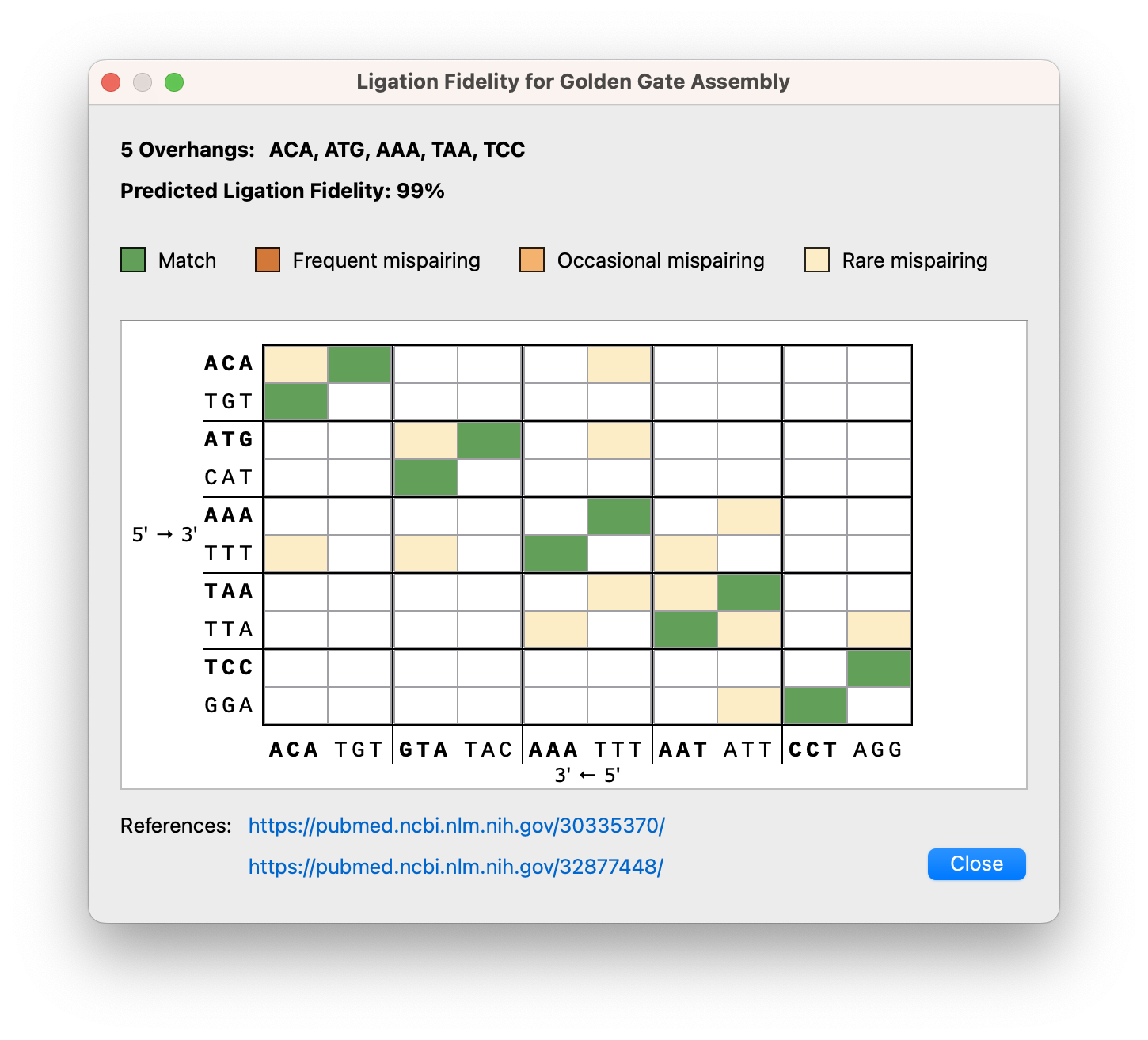
Golden Gate Assembly Improvements
Various enhancements to the Golden Gate cloning dialogs include a ligation fidelity matrix for assessing overall reaction fidelity, it is now possible to adjust overhangs for manually designed primers, a new option to cut the vector with any enzymes (not just Type IIS enzymes), as well as an updated settings dialog with easy access to recommended enzymes.
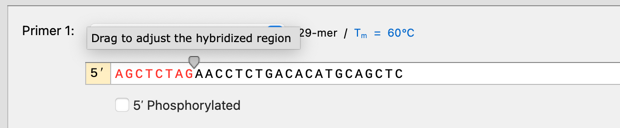
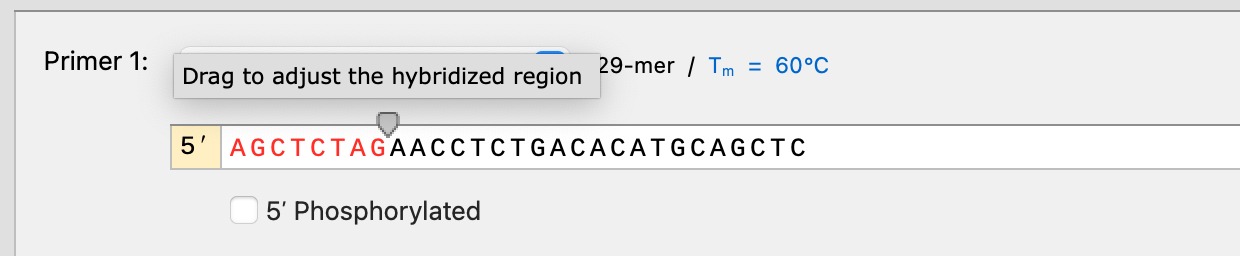
Improvements to primer design in PCR cloning simulations
You can now adjust the hybridization region for all PCR cloning simulations (e.g. PCR, Golden Gate, Gibson, In Fusion, etc.). When using automatic primer design the hybridized region will be configured automatically, ensuring miscellaneous features are not transferred to the product when a 5’ primer extension by chance partially hybridizes to the template.
Protein properties can now be copied and exported
Copy or export the properties and amino acid data for full protein sequences or selected regions in text or spreadsheet (*.csv / *.tsv) formats. Individual properties can also be copied directly from the view.
Modernized toolbar, application, and document icons
SnapGene now uses modernized icons in the top toolbar. The application icon and file icons have also been updated.
This release includes stability improvements as well as fixes for cloning and viewing sequences.
Version 6.1 simplifies primer design when simulating Golden Gate Assembly, includes a new Secondary Structure view for ssRNA sequences, and supports Dark mode across all operating systems.
Golden Gate Assembly
Confidently simulate Golden Gate Assembly with a new tool which can automatically design primers and overhangs for Golden Gate, optimizing the reaction fidelity to maximize the likelihood of success.
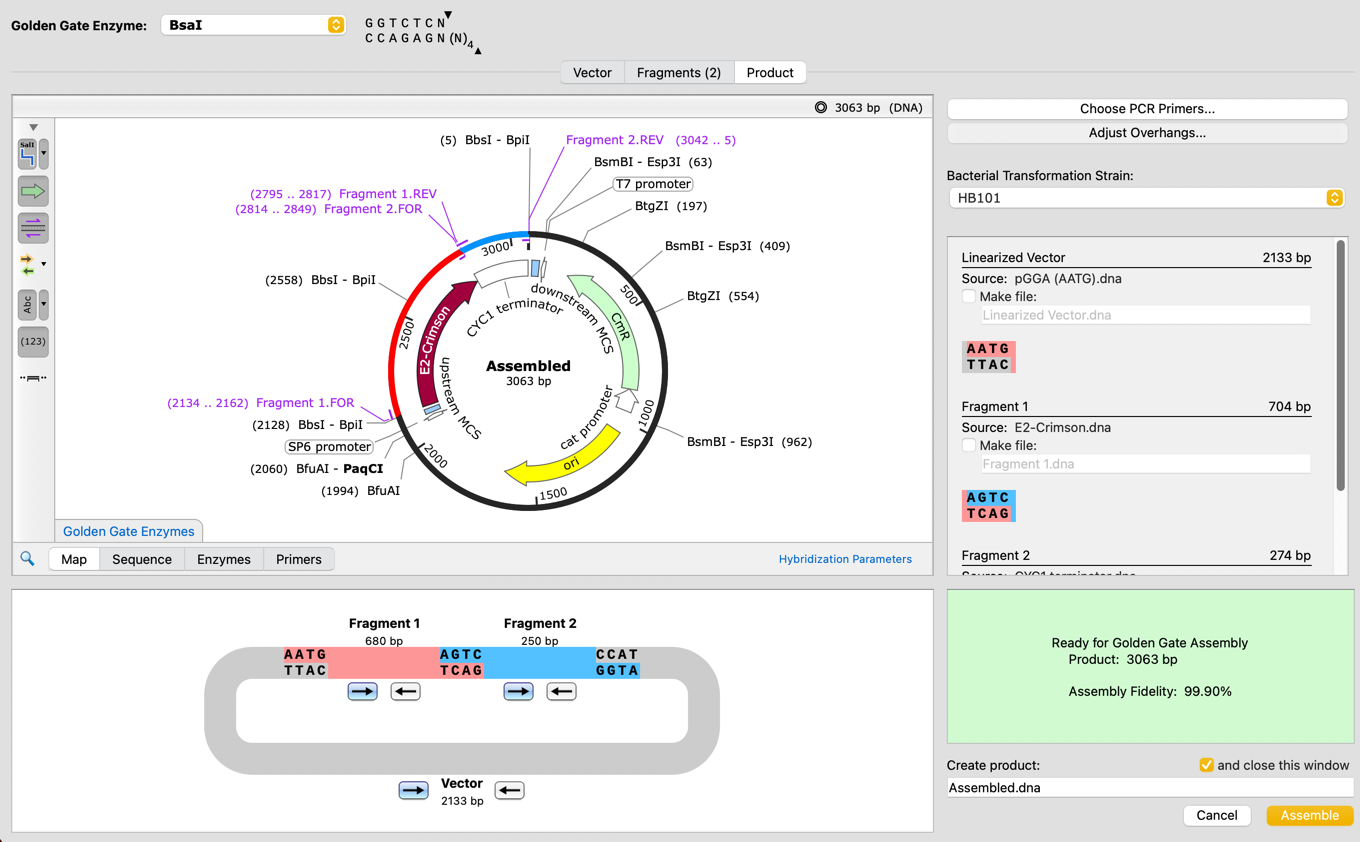
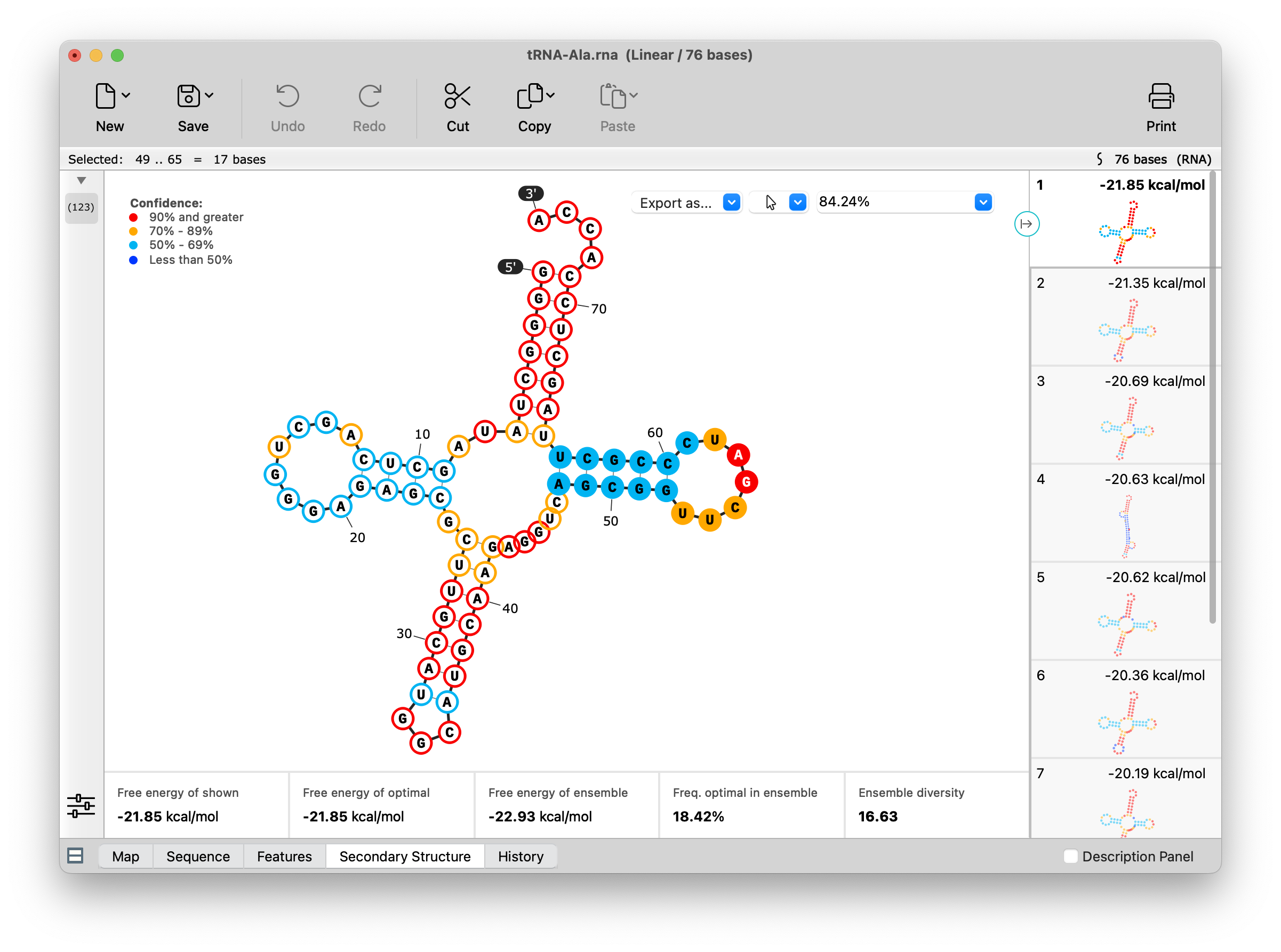
RNA Secondary Structure
Visualize how your single stranded RNA sequences will fold with a new Secondary Structure view which displays the optimal structure as calculated by the ViennaRNA.
Dark Mode
The SnapGene user interface now supports dark mode and by default will match your Dark or Light mode operating system setting.
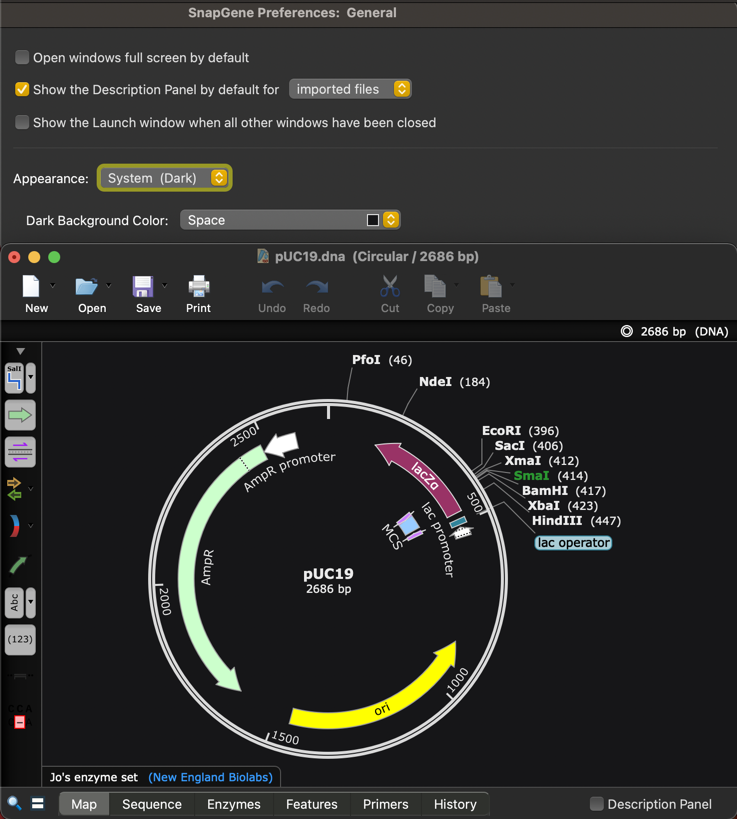
Export Options
Customize how contents is exported to GenBank including LOCUS field identifier feature export options using the new Export pane in Preferences.
Support for Apple Silicon
SnapGene now runs natively on Apple computers with M1 silicon chips.
This release includes an important fix for opening older files.
This release includes important fixes including improved repairing of history in existing files.
This release includes a fix for a serious regression in version 6.0.3 that can result in history not being displayed correctly and ancestral sequences no longer being resurrectable.
This release includes a number of important stability and memory leak fixes and other optimizations.